Two years ago I was asked to participate in the European DNA day essay contest, which every year poses two questions to expand on and selects 3 winners among hundreds of essays from 19 European countries. To my utter bewilderment, I won (second place), and my photo is featured at their site (
http://www.dnaday.eu/winners2015.0.html).
Even though two years have passed, I still read it and ask myself how I was able to write such a good essay (I didn't have any background on the topic, I had just started learning about what genetics were and it was the first time in my life I had to research scientific papers).
As I believe the topic is interesting and not many people know about I thought I would share it here:
How can you explain human complexity when we have so few protein coding genes, e.g. about 5000 less than a cucumber?

Despite humans being considered the most complex organisms on Earth in terms of mental cognition and anatomy, many other species outrank humans in genome size. The marbled lungfish,
Protopterus aethiopicus, has the largest genome with around 132.83 million base pairs, more than forty times the amount of DNA per cell than humans[1]. The same incongruity appears when analysing the number of protein-coding genes of different species. Rice, for instance, has around thirty thousand more genes than humans, and the protozoa
Trichomonas vaginalis around forty thousand more[2]. In fact, biological complexity depends on genomic programming rather than genomic size or the amount of protein-coding genes. This is achieved through gene regulation, which results in differential gene expression and cell specialization. The activation or repression of certain loci requires a delicate balance which can be controlled at different levels of protein synthesis[3],[4].
Before transcription, DNA is condensed into chromatin and wrapped around clusters of eight histone proteins which form nucleosomes. In euchromatin, nucleosomes are loosely packed due to histone acetylation and therefore, RNA polymerase can easily transcribe it. On the other hand, deacetylation results in heterochromatin, which is normally not expressed since it is wound too tightly for transcription to occur. Likewise, DNA methylation adds methyl groups to certain bases, binding chromatin tightly and inhibiting transcription[5].

Two other closely linked processes, imprinting and x-inactivation, play a role in gene expression. Although we receive two copies of each autosomal gene, one from our mother and one from our father, if one of them was epigenetically imprinted during the ovum or sperm formation, it will usually be silenced throughout the organism's whole life[6],[7],[8]. X-inactivation also leads to monoallelic expression as one of the X chromosomes in a female is randomly silenced during early embryonic development. This is done to prevent a potentially critical imbalance of gene expression from a double dose of X-linked genes[8],[9].
The aforementioned methods of regulation are part of what is known as epigenetics: ''the study of heritable changes in gene expression caused by non-genetic mechanisms, thus by alterations other than in the DNA sequence"[10].
However, there is more to DNA than protein-coding genes. Since the 1960's, the parts that are not transcribed into proteins have been referred to as ''junk DNA''. Nevertheless, it is now clear that far from being evolutionary waste, they are the basis for biological complexity. Although the ratio of coding to non-coding genes (ncDNA) differs between species, studies have shown that as biological complexity increases, so does the proportion of ncDNA. While prokaryotes contain about 6% to 14% ncDNA, in humans, it accounts for 98% of the total genome[11],[12].
 |
| Photo cred: The Guardian |
These sequences not only are essential to transcription and translation[13], but also enable the coding of thousands of proteins from a single gene through splicing. Alternative splicing consists of the removal of introns and subsequent joining of exons from a single gene. Depending on how the exons are connected back, different messenger RNA (mRNA) molecules will be produced, each coding for an isoform. Similarly, trans-splicing assembles exons from different pre-mRNAs[14].
 |
| Photo cred: Gostica |
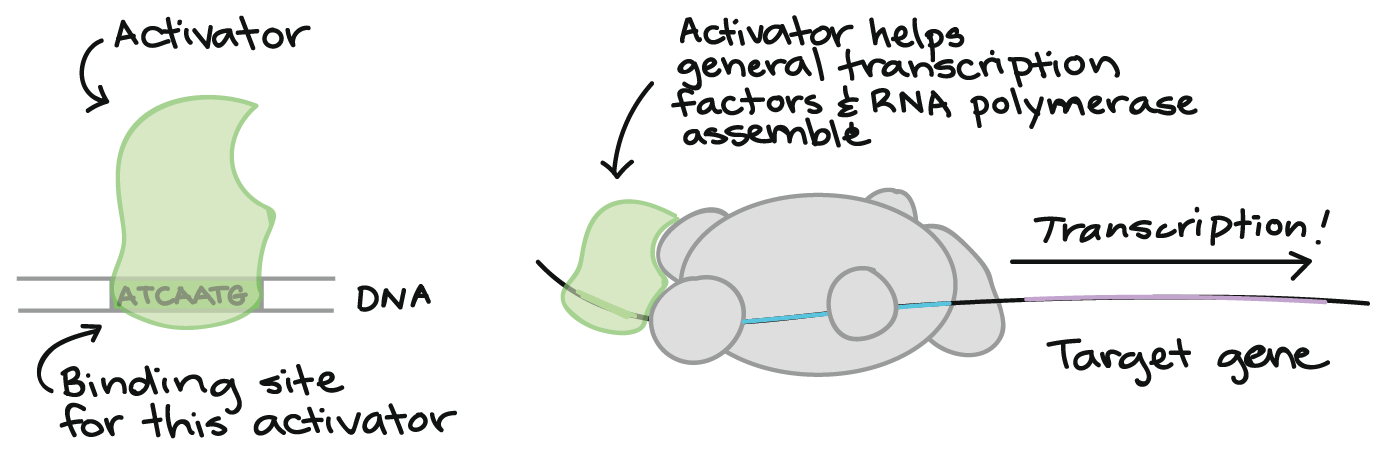
Furthermore, transcription factors (TFs), proteins that inhibit or facilitate gene transcription, need to bind to cis-regulatory elements, sequences of ncRNA that include promoters and enhacers[15],[16]. Moreover, microRNA and small inhibitory RNA (siRNA) hinder the translation of a targeted mRNA by winding round it. While siRNAs only silence the locus from which they originated, microRNAs are estimated to influence the translation of one third of all genes in mammals managing cell differentiation and growth, mobility and apoptosis[13],[17].
 |
| Photo cred: Khan Academy |
 |
| Photo cred: Khan Academy |
 |
| Photo cred: longevinex |
 |
| Photo cred: Currents in Biology |
In addition, transposable elements (TEs), also known as "jumping genes" or transposons, are thought to be another means of regulation. These sequences of DNA, first described by Barbara McClintock in the 1940's[18], can change their position within the genome, creating or reversing mutations[19],[20].
Other mechanisms include RNA editing, in which the sequence of a mRNA is modified resulting in the codification of a different protein; and tandem chimerism, by which two adjacent genes are transcribed into a single mRNA[3],[21],[22].
Ergo, biological complexity derives from the aforementioned molecular systems, which allow higher eukaryotes to carry out a plethora of genetic responses to their environment. All these processes in charge of controlling cell specialization become increasingly intricate as so does the organism's complexity. Although humans merely have 20,000 protein-coding genes, our incredibly elaborate repertoire of gene regulation mechanisms makes the codification of about 500,000 or more distinct human proteins possible[2].
References
[1] Gregory, T.R. (2005). Animal Genome Size Database. http://www.genomesize.com.
[2] Pray, L. (2008) Eukaryotic genome complexity. Nature Education 1(1):96
[3] Hoopes, L. (2008) Introduction to the gene expression
and regulation topic room. Nature Education
1(1):160
[4] Klug, W., & Cummings, M. (1999). Conceptos de
genética. Madrid: Prentice Hall Iberia.
[5]Epigenetics: Fundamentals. (n.d.).
http://www.whatisepigenetics.com/fundamentals/
[6] Genomic Imprinting. (n.d.).
http://learn.genetics.utah.edu/content/epigenetics/imprinting/
[7] Jirtle, R. (n.d.). Geneimprint.
http://www.geneimprint.com/
[8] Phillips, T. & Lobo, I. (2008) Genetic imprinting
and X inactivation. Nature Education
1(1):117
[9] Ahn, J. & Lee, J. (2008) X chromosome: X
inactivation. Nature Education
1(1):24
[10] Brouwer J.R. (2012, April 4). A Crash Course in
Epigenetics Part 1: An intro to epigenetics. Bitesize Bio.
[11] Taft, R. & Mattick, J. (n.d.) Increasing Biological
Complexity Is Positively Correlated with the Relative Genome-wide Expansion of
Non-protein-coding DNA Sequences. http://arxiv.org/pdf/q-bio/0401020
[12]Mandal, A. (2010, May 7) What is Junk DNA?
http://www.news-medical.net/health/What-is-Junk-DNA.aspx
[13] Clancy, S. (2008) RNA functions. Nature Education 1(1):102
[14] Clancy, S. (2008) RNA splicing: introns, exons and
spliceosome. Nature Education 1(1):31
[15] Phillips, T. (2008) Regulation of transcription and
gene expression in eukaryotes. Nature
Education 1(1):199
[16] Phillips, T. & Hoopes, L. (2008) Transcription
factors and transcriptional control in eukaryotic cells. Nature Education 1(1):119
[17] Meštrović, T. (2010, February 3) What is MicroRNA? http://www.news-medical.net/health/What-is-MicroRNA.aspx
[18] Pray, L. & Zhaurova, K. (2008) Barbara McClintock
and the discovery of jumping genes (transposons). Nature Education 1(1):169
[19] Pray, L. (2008) Transposons, or jumping genes: Not junk
DNA? Nature Education 1(1):32
[20] Moalem, S., & Prince, J. (2007). Survival of the
sickest: A medical maverick discovers why we need disease. New York: William
Morrow.
[21] Simpson, L.
& Emerson RB. (1996) RNA Editing. http://www.ncbi.nlm.nih.gov/pubmed/8833435RNA
Editing. Annu Rev Neurosci.
[22]Parra, G. (2005).
Tandem chimerism as a means to increase protein complexity in the human genome.
Genome Research, 37-44.









